Introduction
Core laboratories must balance the efficiency of their NGS workflows with the quality of the data they generate when providing a variety of next-generation sequencing (NGS) services related to a wide range of clients, sample types, and sequencing methods. As the cost of sequencing decreases, the costs associated with library preparation are starting to become a large part of the overall NGS workflow costs. To address this major cost issue in sequencing, many institutions have followed the trend of reaction miniaturization, effectively reducing reaction volumes to the manufacturer's recommended volume of 10%.
This application note highlights the workflow for generating high-quality NGS libraries with precise positive displacement microfluidic reagent dispensing even in the presence of low cell input. Quality control data from a quarter-volume Illumina Nextera XT library preparation demonstrates the applicability and effectiveness of reaction miniaturization in the context of single-cell sequencing.
Materials .
- Eppendorf twin.tec 384 plate (PN 0030129342)
- Illumina Nextera XT DNA Library Preparation Kit (PN FC-131-1096)
- Illumina Nextera XT Indexing Kit V2 (FC-131-2001)
- Thermal Cycler
- FORMULATRIX® MANTIS® Liquid Processor (PN MANTV3.2)
- Alpaqua 384 Column Magnetic Plate (PN A001222)
- ThermoFisher Qubit 4 Fluorometer (PN Q33226)
- Agilent 2100 Bioanalyzer (PN G2939BA)
Methods
Quarter-volume Illumina Nextera XT Library Preparation
Eight mouse neuronal cell cDNA samples (3 cell types; 1 ng of input DNA) were used in this pilot project, and then the quality of the library generated in one-quarter volume was assessed by the following experimental protocol:
**A. Tagment cDNA - total reaction volume of 6.35 µL **
Add 1.25 µL input DNA to the wells of a 384-well microtiter plate.
Draw TD Buffer into a 200 µL pipette tip* and place it on the LV chip of the MANTIS. Dispense 2.5 µL of TD Buffer into each well containing a sample on the MANTIS microplate.
Mix.
Pipette the ATM into a 200 µL pipette tip* and place it on the LV chip of the MANTIS. Dispense 1.3 µL of ATM into each well of the MANTIS microplate containing the sample.
Mix.
Thermal cycling.
i. Hold at 55°C for 5 minutes
ii. after that it was kept at 10°C
Draw NT buffer into a 200 µL pipette tip* and place it on the LV chip of the MANTIS. Dispense 1.3 µL of NT buffer into each well containing samples on the MANTIS microplate.
Mix.
Incubate at room temperature for 5 minutes.
B. Library amplification - 12.65 µL total reaction volume
Pipette NPM into a 200 µL pipette tip* and place on the LV chip of the MANTIS. Dispense 3.8 µL into each well containing the sample on the MANTIS microplate.
Manually pipette 1.25 µL of both index primers into the samples to obtain a unique combination of index primers for each sample.
Mix.
Thermal cycling.
i. 3 minutes at 72°C
ii. 30 seconds at 95°C
iii. do the following 12 cycles
a. 10 seconds at 95°C
b. 30 seconds at 55°C
c. 30 seconds at 72°C
iv. 5 minutes at 72°C
v. Maintain 10°C at all times
C. Library cleanup
1. Draw AMpure XP microbeads into a 200 µL pipette tip* and place on the HV chip of the MANTIS. Dispense 10 µL into each well of the MANTIS microplate containing the sample.
2. Mixing.
3. Incubate at room temperature for 5 minutes
4. Place the microplate on the magnet and wait 2 minutes
5. Manually remove all supernatant (do not remove the microplate from the magnet until step 11)
6. Using the Continuous Flow function option on the MANTIS, add 50 µL of 80% EtOH to each well containing the sample on the microplate.
7. Incubate for 30 seconds.
8. Manually remove all supernatant while placing the microplate on the magnet.
9. Repeat steps 6-8. 1. Air dry for 15 minutes. 2. 2. Remove the microplate from the magnet. 3. 3. Pipette RSB into a 200 µL pipette tip* and place on the HV chip on the MANTIS. 4. Dispense 15 µL into each well of the MANTIS microplate containing the sample. 5. Mix. 6. 6. Incubate at room temperature for 2 minutes. 7. 7. Place the microplate on the magnet. 8. 8. Incubate for 2 minutes. 9. 9. Manually transfer 12.5 µL from the microbeads to the new microplate.
D. Aspirate volume = Dispense volume per sample x number of samples + 10% safety factor. Use 1000 µL pipette tips when aspiration volumes are greater than 200 µL.
Results.
Nextera library quality is assessed for each sample by tracking with Agilent BioAnalyzer.
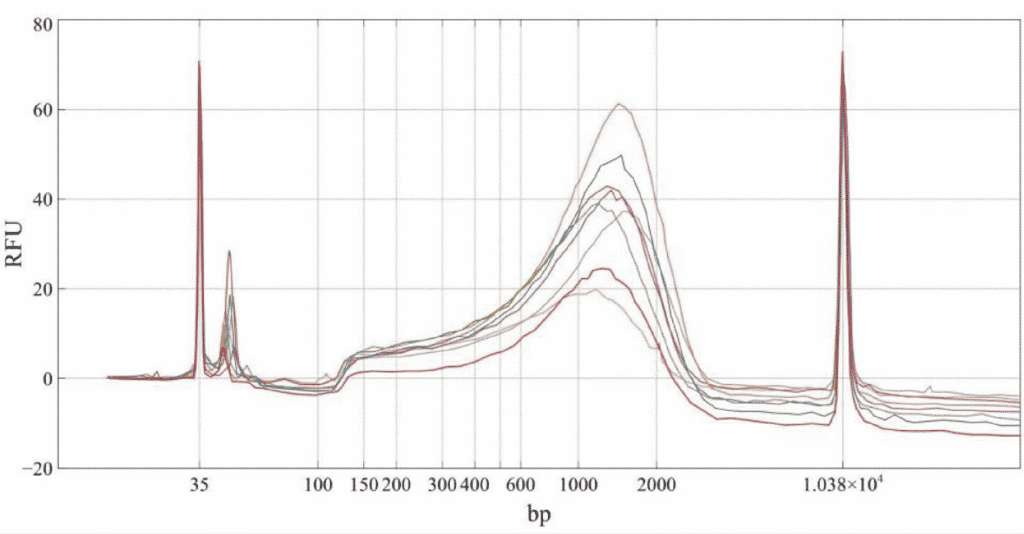
Figure 1. Bio nalyzer tracing of 8 next-generation sequencing libraries prepared from 8 unique 1ng murine neuronal cell cDNA samples
Summary.
According to BioAnalyzer data tracking, the library composition prepared by quarter-volume reaction miniaturization is acceptable and comparable to that prepared according to the full-volume experimental protocol.
Preparing libraries in one-quarter volumes provides significant savings of 75% per reaction kit cost. FORMULATRIX®'s MANTIS® Liquid Processor facilitates reaction miniaturization by performing precise microfluidic reagent dispensing in volumes as low as 100 nL, and is consumable-free, simple and reliable to use.
Source: @BostonFummerle
Time: February 10, 2022
